Primate Phylogeny
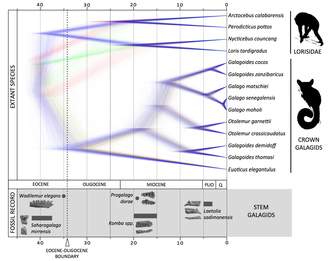
Evolutionary History of lorisiform primates
|
Background. Lorisiformes (Lorisidae and Galagidae) are among the most morphologically cryptic of all primates and their diversity and relationships are some of the most longstanding problems in primatology. Our knowledge of their evolutionary history has been limited by a lack of appropriate molecular data and a paucity of fossils. Most phylogenetic studies have produced conflicting results for many clades, and even the relationships among genera remain uncertain.
Goals. To clarify lorisoid evolutionary history by combining genetic and morphological data. |
Publications:
- Pozzi L. (2016). The role of forest expansion and contraction in species diversification among galagos (Primates: Galagidae). Journal of Biogeography, 43: 1930–1941
- Pozzi L., Nekaris K.A.I., Perkin A., Bearder S.K., Pimley E.R., Schulze H., Streicher U., Nadler, T., Kitchener A., Zischler H., Zinner D., Roos C. (2015). Remarkable Ancient Divergences Amongst Neglected Lorisiform Primates. Zoological Journal of the Linnean Society, 175: 661-674.
- Pozzi L., Disotell T.R., Masters J.C. (2014). A multilocus phylogeny reveals deep lineages within African galagids (Primates: Galagidae). BMC Evolutionary Biology 2014 14:72.
- Masters J.C., Boniotto M., Crovella S., Roos C., Pozzi L., DelPero M. (2007). Phylogenetic relationships among the lorisoidea as indicated by craniodental morphology and mitochondrial sequence data. Am J Primatol 69:6-15.
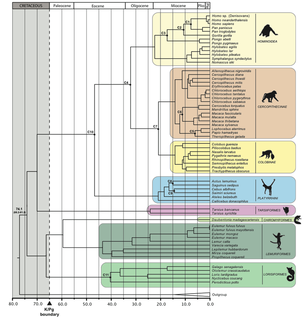
Primate origins and integration of fossil data
|
Background. Understanding the origins of the order Primates is, by its own nature, one of the major questions in primatology. Despite the great interest in this topic, a consensus about the time of the primate radiation has remained elusive because of the contrast between the fossil record and molecular estimates. In particular, molecular estimates have tended to yield older dates, in some cases much older than has seemed credible based on the paleanthropological record.
Goals. To reconstruct phylogenetic relationships and to estimate divergence times within primates using complete mitochondrial genomes. In this project, we also explored the importance of calibration points in estimating time using molecular data. We focused particularly on identifying inconsistent fossils when multiple fossil calibrations are available for a clade. |
Publications:
- Pozzi L., Hodgson J.A, Burrell A.S., Sterner K.N., Raaum R.L., Disotell T.R. (2014). Primate phylogenetic relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol, 75:165-183.
- Pozzi L., Hodgson, J.A., Burrell A.S., Disotell, T.R. (2011). The stem Catarrhine Saadanius does not inform the timing of the origin of crown Catarrhines. J Hum Evol, 61:209-210
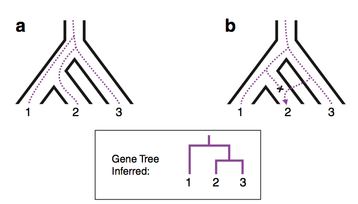
Gene tree - Species tree
|
The concatenation of multiple loci has been used extensively in primatology although, despite the practical advantages of this approach, both simulation and empirical studies have shown that this method can perform poorly in cases of high tree discordance across different loci. More specifically, phylogenetic reconstructions based on concatenated datasets do not account for individual gene histories, and can therefore produce misleading topologies, with most of the nodes highly supported (high bootstrap values and/or posterior probabilities) despite their not reflecting the actual evolutionary history of the species.
|
Over the last few years I have been exploring the use of gene tree-species tree methods in reconstructing primate phylogeny. These methods use a coalescence approach that takes into account the possible discordance among genes – mainly as a consequence of incomplete lineage sorting (ILS) – and reconstructs the species tree within which each individual gene tree is embedded. My main goal is to explore the level of tree discordance in different primate lineages and to better understand pros and cons of coalescence methods in reconstructing phylogenies. Coalescence methods can provide the first step to further investigate the processes that caused tree incongruence across different regions of the genome.
Publications:
- Pozzi L., Disotell T.R., Masters J.C. (2014). A multilocus phylogeny reveals deep lineages within African galagids (Primates: Galagidae). BMC Evolutionary Biology 2014 14:72.
- Pozzi L., Bergey, C.M., Burrell A.S. (2014). The use (and misuse) of phylogenetic trees in comparative behavioral analyses. Int J Primatol, 35:32-54.
Other projects in primate phylogeny
- Molecular phylogeny of lemurs [DelPero et al. 2006]
- Phylogeny and biogeography of squirrel monkeys (Saimiri spp.) [Chou et al., 2011]